Single-cell analysis techniques such as fluorescence-activated cell sorting (FACS) and single-cell DNA and RNA sequencing have broadened researchers’ understanding of cell biology in health and disease by enabling the study of rare cell types and phenotype variations within same-cell populations. However, new methods are evolving rapidly, driven by the need for molecular profiling of individual cells within their native spatial context. Here, we look at some of these so-called spatial-omics technologies, including key protocol differences. We also share insights from Lindsy Rapkin, PhD, Field Applications Scientist at Standard BioTools™, who explains how Imaging Mass Cytometry™ (IMC™) has been combined with RNAscope™ in situ hybridization (ISH) technology to simultaneously visualize key protein and RNA markers in tissue samples.
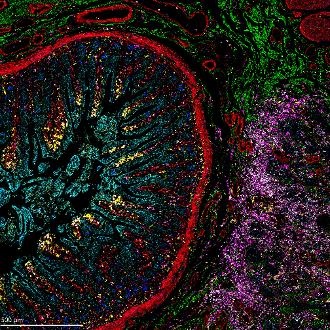
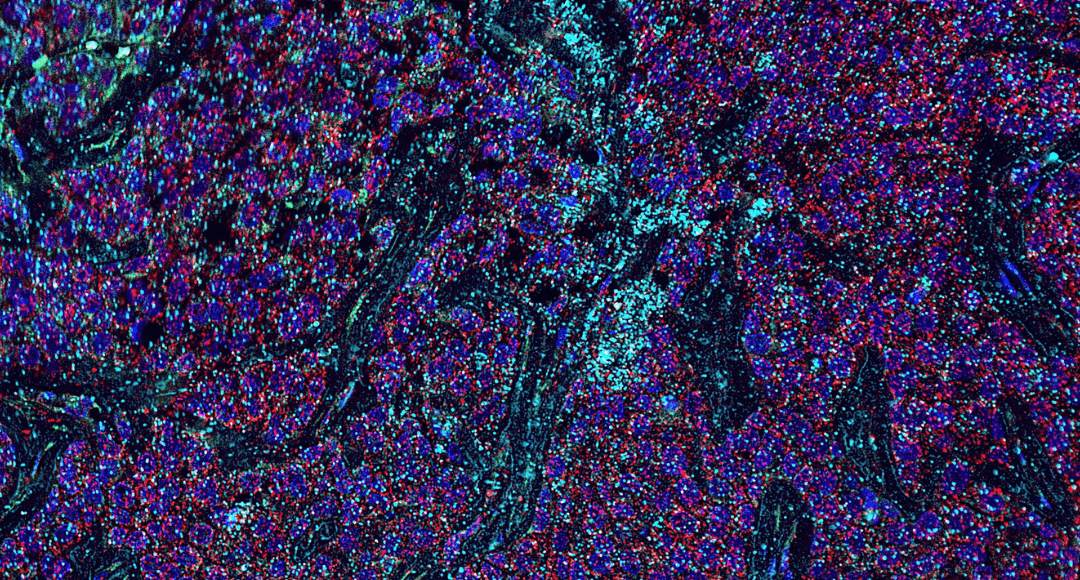
Figure 1. IMC staining of pancreatic ductal adenocarcinoma (PDAC) tissue showing 7 markers: aSMA (red), HLADR (yellow), E-cadherin (cyan), collagen 1 (green), CD68 (magenta), INOS (blue) and Granzyme B (white).
What is Spatial-Omics?
Omics refers to a broad range of technologies used for characterizing different biomolecules in cells or tissues. Specifically, genomics technologies allow for studying the whole genome of a cell or organism; epigenomics technologies are used for investigating chemical modifications to DNA, which can alter gene expression; and transcriptomics technologies enable the analysis of RNA transcripts. Proteomics and metabolomics technologies are respectively used for characterizing proteins and metabolites. Spatial-omics is the application of one or more of these technologies in situ, typically using formalin-fixed, paraffin-embedded (FFPE) tissue samples. Bringing spatial context to omics data is important since the functional state of any given cell can be modulated by its neighbors, either through direct interaction, via signaling molecules or by microenvironmental factors such as chemical compound gradients.
Spatial-Omics Technologies
Established methods for spatial profiling include immunohistochemistry (IHC), which uses fluorophore-labeled antibodies for detecting proteins of interest, and fluorescence in situ hybridization (FISH), which relies on fluorescent probes to bind specific DNA target sequences. Yet, while these techniques still have broad applicability for scientific research, they are now complemented by a vast array of more than 50 advanced technologies that have been developed to overcome known limitations of existing methods1. With so many different options available for spatial profiling, it is impossible to cover every technology here. As such, the following are some of the main approaches on which these newer modalities are based:
Cyclical IHC with Fluorophore-Labeled Antibodies
A major drawback of traditional immunofluorescent IHC is that it allows for detecting only a handful of protein markers due to spectral overlap. However, it has been found that the multiplexing capacity of IHC can be increased by performing sequential rounds of staining, imaging, and stripping/bleaching. Methods based on this strategy include cyclic immunofluorescence (cycIF), which involves mild chemical inactivation of common dyes after each image acquisition; iterative bleaching extends multiplexity (IBEX), which rapidly bleaches fluorophores without causing epitope loss or tissue destruction; and MACSima™ Imaging Cyclic Staining (MICS), which incorporates both photobleaching and the controlled release of antibodies (REAlease® Antibodies) or their labels (REAdye_lease™ Antibodies)2,3,4. These techniques allow for detecting tens to hundreds of different protein targets in the same tissue sample but require careful implementation to avoid compromising antibody binding.
Use of DNA-Barcoded Antibodies
Like fluorophore-labeled antibodies, DNA-barcoded antibodies can be employed for detecting proteins of interest, typically through their use with fluorescently labeled complementary DNA probes. A technology based on this approach known as immunostaining with signal amplification by exchange reaction (Immuno-SABER) has been shown to achieve up to 180-fold signal amplification using standard detection equipment5. Another method termed co-detection by indexing (CODEX) involves iterative cycles of staining and imaging and is claimed to have unlimited multiplexing capacity6.
mRNA Probes for Targeted Transcripts
mRNA probes have been widely adopted for spatial transcriptomics, with some methods now able to detect up to 10,000 transcripts. This high multiplexing is achieved using a combination of spectral barcoding, whereby different fluorophores are paired with specific mRNA target sequences, and temporal barcoding, which is based on multiple rounds of probe hybridization and stripping1. Examples of spatial transcriptomics technologies developed within the last decade include multiplexed error robust fluorescence in situ hybridization (MERFISH), sequential fluorescence in situ hybridization (seqFISH), and Multi Omic Single-scan Assay with Integrated Combinatorial Analysis (MOSAICA). These serve as popular alternatives to the established method of performing laser capture microdissection (LCM) followed by single-cell RNA sequencing (scRNA-seq), which has a complex and time-consuming workflow.
Imaging Mass Cytometry
Imaging Mass Cytometry, based on CyTOF® technology, uses antibodies labeled with metal tags for spatial proteomics. “Once the sample has been immunostained, it is introduced into a Hyperion XTi™ Imaging System, where it is ablated by a UV laser, one pixel at a time, before being transferred to an inductively coupled plasma for ionization,” Rapkin explains. “The ions are then quantified and the signals corresponding to each tag are correlated back to the respective markers.” A major advantage of using metal tags instead of fluorophores is that there is no need to worry about spectral overlap or autofluorescence. As a result, IMC allows for imaging as many as 40 or more markers simultaneously, making it more efficient than cyclical IHC methods.
Multimodal Spatial Profiling with IMC and RNAscope
Standard BioTools has developed a simple three-step workflow for co-detecting proteins and RNA with a combination of IMC and RNAscope ISH Technology (from Advanced Cell Diagnostics, a Bio-Techne brand). First, oligonucleotides are labeled for RNAscope. The RNAscope assay is then performed, and the samples are incubated overnight with IMC antibodies. A standard image analysis pipeline is used for deciphering the data.
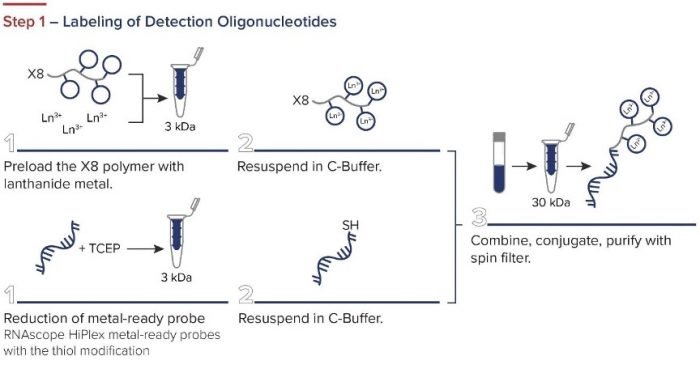
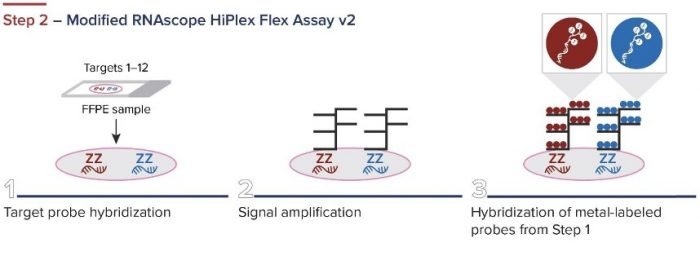
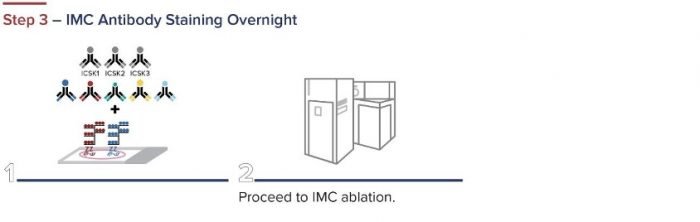
Figure 2. The integrated IMC and RNAscope workflow.
“While detecting protein targets allows for determining cellular identity, interrogating the transcriptome can provide a deeper understanding of cellular function and activation state,” Rapkin says. “By applying IMC and RNAscope for spatial profiling within the tumor microenvironment, we have demonstrated the value of multimodal spatial profiling for investigating tumor-immune interactions. This strategy could readily be applied across other fields of research for deeper insights into sample material.” To learn more, visit https://www.standardbio.com/resources/blog-articles/2023/06/imc-rnascope-poster.
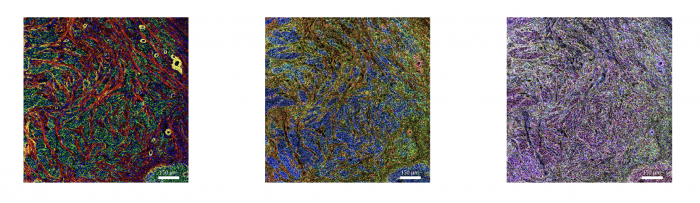
Figure 3. IMC and RNAscope co-detection of a breast cancer sample. IMC (left panel) showing the structural markers Collagen 1 (red), E-cadherin (green), and aSMA (yellow), plus DNA (blue). RNAscope (middle panel) showing UBC (red), B2M (green), and GAPDH (blue). Maxpar IMC Cell Segmentation kit staining (right panel).
Technologies for Epigenomics and Metabolomics
Although many different technologies have been developed for spatial proteomics and transcriptomics, fewer methods exist for spatial epigenomics and metabolomics. Available options for epigenomics research include spatial-ATAC-seq, which provides spatially resolved chromatin accessibility profiling via next-generation sequencing, and Spatial-CUT&Tag, which delivers genome-wide profiling of histone modifications7,8. For metabolomics, metaFISH pairs FISH with matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI-IMS) to map metabolic species in native tissues, while airflow-assisted desorption electrospray ionization (AFADESI)-IMS provides untargeted analysis of more than 1,500 metabolites9,10.
Supporting Your Research
As spatial-omics technologies evolve, tools that streamline panel design and ensure experimental success are more important than ever. FluoroFinder supports researchers with user-friendly, cloud-based tools designed for high-parameter experiments. Our Panel Builder helps you efficiently design and visualize complex panels for techniques like IMC™, FACS, and fluorescent imaging, while our Antibody Search platform aggregates validated antibody data from all suppliers, enabling quick comparison of clones and conjugates for multiplex studies. Whether you’re planning a spatial transcriptomics experiment or integrating protein and RNA detection using IMC and RNAscope™, FluoroFinder’s tools can help you make informed decisions and accelerate discovery.