Cellular function encompasses the basic processes a cell must undertake to do it’s assigned job. That could include protein production, enzymatic degradation, proliferation, apoptosis, cell signaling, mitochondrial respiration, buffering reactive oxygen species, etc, any task that is required to maintain the health and equilibrium of the organism. Thus, any assay that assesses the state of any of these processes is called a functional probe whether it is chromogenic, fluorescent or luminescent. Here are a few assays that are particularly well suited to live-cell imaging in real time and over a time course.
Calcium Imaging and Ion Indicators
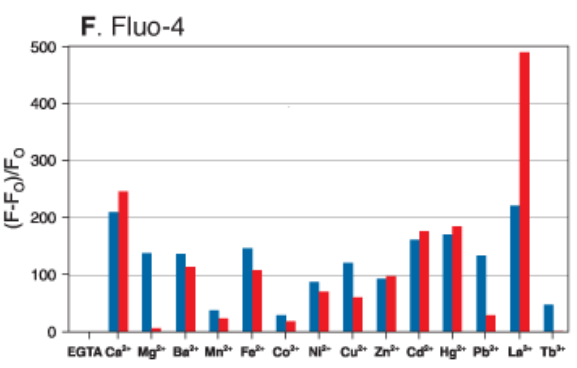
Action potentials are regulated by calcium, sodium and potassium channels working in concert to propagate signaling and maintain homeostasis in muscles and the nervous system. Cellular probes called ion indicators will have an affinity (Kd) for a particular ion, however they are never specific for that ion. The most popular single wavelength (one excitation peak, one emission peak that becomes brighter when bound) calcium indicator is Fluo-4 developed in the lab of the late, great Roger Tsien. The graph below is taken from the Molecular Probes handbook showing the relative intensity change for 1uM (blue bar) and 100uM (red) ion concentrations. The element that elicits the strongest response from Fluo-4 is actually Lanthanum. Luckily for us, there shouldn’t be any lanthanum in a healthy human biological sample. Second to that is calcium. This graph can also be misleading since the element most likely to cause competition for calcium in a biological sample is actually magnesium, which was assayed at a 100x higher concentration to calcium although they are plotted together on the same scale. So, we say these indicators are selective, not specific. Also, because ion indicators chelate the ion they are sensing, it will deplete that ion from bioavailability in the sample, which can be harmful to cell health in long-term experiments.
Ion indicators can come in a few varieties. The salt form is not cell-permeant and can be used in microplate assays to quantitate ionic concentration in cell extracts or solutions. The acetoxymethylester (-AM) versions are passively cell-permeant to mammalian cells and retained after the -AM moiety is hydrolyzed by intracellular esterase. Some indicators can also come in versions that can be covalently conjugated to proteins or other biomolecules to target the indicator to a particular location within the cell or tissue. Ion indication is fluorogenic which means the probe emits like an on-off switch, on when bound, off when unbound. However, the binding affinity varies quite a bit between different indicators and their ions. Some have a quick off rate and quick recovery time, helpful in calcium imaging for measuring accurate timing of an action potential pulse.
There are also ratiometric probes where either two excitation peaks or two emission peaks are monitored in relationship to ion binding. This can often be more sensitive to subtle changes than a single-wavelength indicator. In addition to chemical probes there are fluorescent proteins that have been developed for more stable neuronal imaging of calcium flux called GCaMP as described by Zhang et al in a recent Nature publication (1). In this paper, they employ multiphoton live-cell imaging of specific neurons expressing GCaMP specifically a variant called jGCaMP8.
Expression Tags
Image Courtesy of Abberior
Live-cell imaging has benefited from the advances made in the diversity of fluorescent proteins, which allow us to monitor localization and expression levels of proteins in real time. There are unique alternatives to traditional genomically encoded inherently fluorescent proteins like GFP. Alternatives like Halo-Tag from Promega or the SNAP-tag and CLIP-tag from New England Biolab are also expression tags albeit not inherently fluorescent ones. They are also commonly used in a transient manner rather than genomically encoded and a strong advantage is their smaller size as compared to GFP derivatives. The proteins expressing these tags can be visualized by binding an exogenously applied fluorescent ligand. For live-cell imaging, the Si-rhodamine Janelia Fluor dyes and Abberior’s LIVE-SNAP labels are passively cell permeant and are fluorogenic in that they become fluorescent when bound to the expression tag, making them helpful for no-wash methods. An interesting use of these tags is to visualize a time course of newly synthesized protein and their trafficking using different color variants of the fluorescent ligand delivered in a “pulse-chase” method over multiple timepoints. In addition, since SNAP and Halo-tags covalently react to their ligands via different chemical mechanisms, the tags can be expressed on different proteins for simultaneous multicolor labeling at each timepoint. A good example of this method is a paper characterizing mitochondrial dysfunction, protein turnover and degradation from Chantell Evans at Duke and Erika Holzbaur at UPenn (2) .
These are just two classes of live-cell imaging probes out of many that allow us to understand the function of individual cells, their organization and how they maintain health in an organism. There is no singular resource to learn more about chemical probes and how they relate to your research. However, online I could recommend the Molecular Probes handbook hosted by Thermo Fisher and the MicroscopyU website hosted by Nikon. However, excellent courses are also available through the Marine Biological Laboratories found here: https://www.mbl.edu/education/advanced-research-training-courses/course-offerings.
References
- Zhang Y, Rózsa M, Liang Y, et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature. 2023;615(7954):884-891. [doi:10.1038/s41586-023-05828-9] (https://www.nature.com/articles/s41586-023-05828-9)
- vans CS, Holzbaur EL. Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. Elife. 2020;9:e50260. Published 2020 Jan 14. [doi:10.7554/eLife.50260](https://elifesciences.org/articles/50260)